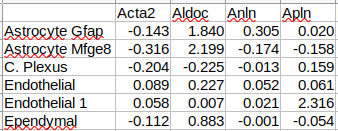
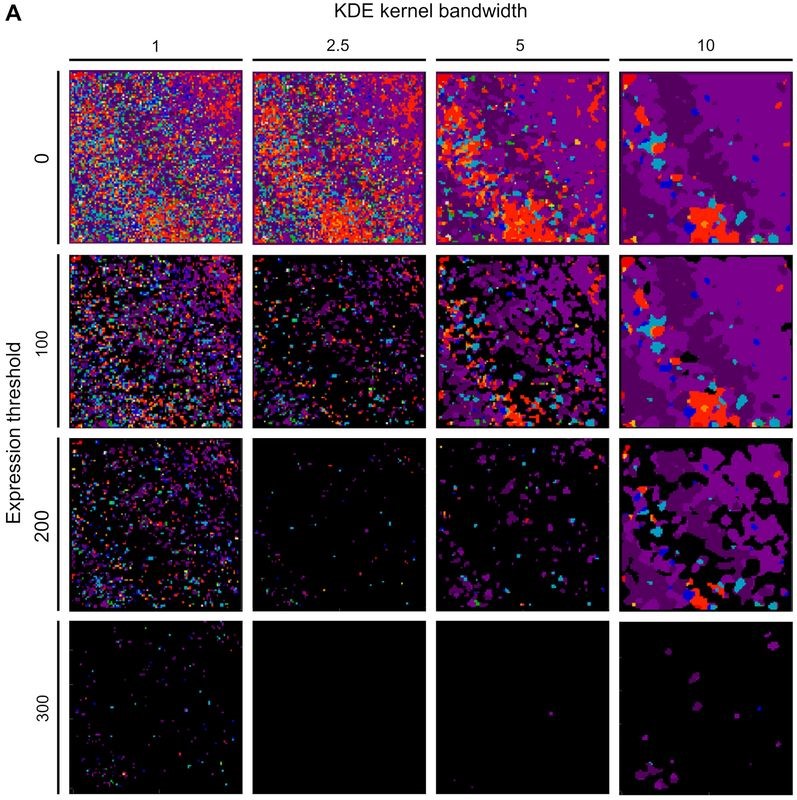
Spatial Sequencing Analysis Pipeline.
SSAM-lite is a complete spatial sequencing analysis pipeline that runs entirely in your browser.
Data Center
Click or drag-and-drop to upload your data.
Parameters
Use previews to find an optimal set of parameters for your analysis.
Use preview generator for parameter search
Analysis
Step 1: Kernel Density Estimation
As a first step, we run a KDE that creates a density of mRNA expression.
Run Kernel Density Estimation
Step 2: Cell Assignments
We can now infer cell types.
Infer Cell Types
Clear analysis
About
The SSAM framework was conceived by Dr. Jeongbin Park
as part of his PhD project at the Berlin Institute of Health.
Read the original SSAM publication
to learn more about the methods.
SSAM-lite has been designed and implemented by Sebastian Tiesmeyer at the Berlin Institute of
Health.
Learn more about SSAM-lite here.
For a comprehensive documentation of the SSAM-lite tool check out our
Read the
Docs page.
If you are using SSAM-lite for your research please cite:
Tiesmeyer S, Sahay S, Müller-Bötticher N, Eils R, Mackowiak SD and Ishaque N (2022)
SSAM-lite: A Light-Weight Web App for Rapid Analysis of Spatially Resolved Transcriptomics Data.
Front. Genet. 13:785877. doi: 10.3389/fgene.2022.785877
SSAM-lite: A Light-Weight Web App for Rapid Analysis of Spatially Resolved Transcriptomics Data.
Front. Genet. 13:785877. doi: 10.3389/fgene.2022.785877